From Concept to Functional Prototype in One Hour Using AI
Using AI-assisted development to build a working 3D protein visualization tool that connects to AlphaFold, renders molecular structures, and identifies potential binding sites.
Project Summary
Role: Designer and builder
Tools: Claude Sonnet 4 (prompt development), Bolt.new (prototyping and build)
Duration: 1 hour from concept to working prototype
Domain: Biotech research tools
The Question
How quickly can a designer with domain knowledge go from an idea to a functional, testable prototype using AI-assisted development?
Traditional prototyping tools such as Figma are effective for screen-level design, but they struggle to model complex interactive workflows, including 3D visualization, real-time data rendering, and API-driven functionality. For specialized domains such as biotechnology, the gap between a static mockup and a system that researchers can interact with can require weeks of design and engineering coordination.
I wanted to test whether AI development tools could dramatically close that gap, using a problem space I know well. My undergraduate research at Loyola (degree in Biology) focused on determining the protein structure of hepatitis B surface antigen, so I brought genuine domain knowledge to the problem, not just design skills.
The Approach
I began with Claude's Sonnet 4 to develop a well-structured prompt, rather than learning Bolt's syntax through trial and error. This prompt-engineering step is itself part of the methodology: using one AI tool to optimize input for another.
Before committing to Bolt, I ran quick tests with Lovable and v0 as alternatives. Bolt produced the best results with the fewest manual fixes.
My initial prompt asked Bolt to create a protein folding viewer that connects to AlphaFold's database and renders the UI in dark mode. From there, I forked the project and extended it to identify potential binding sites and display their relative quality, a feature that would be directly useful to researchers evaluating drug targets.
What it Does
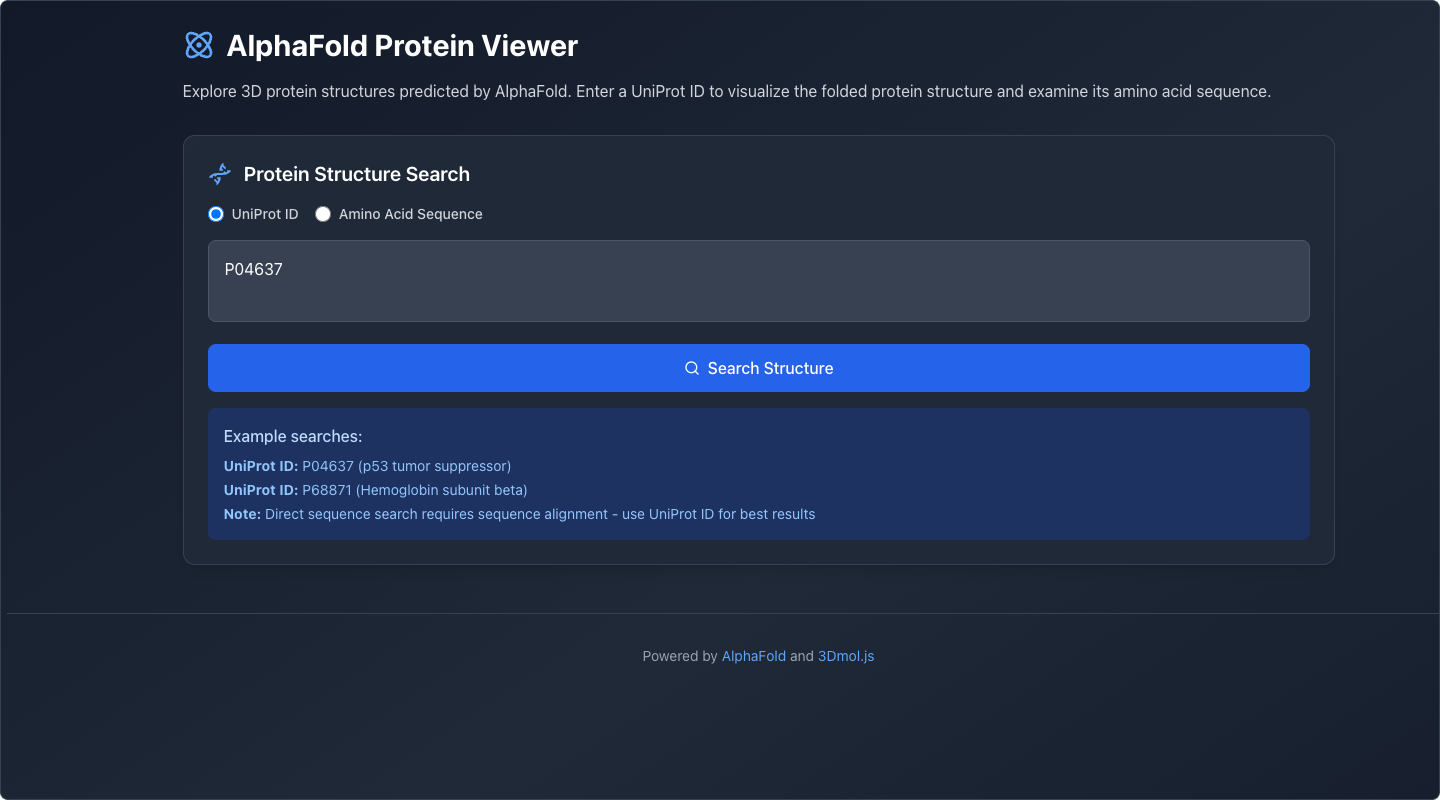
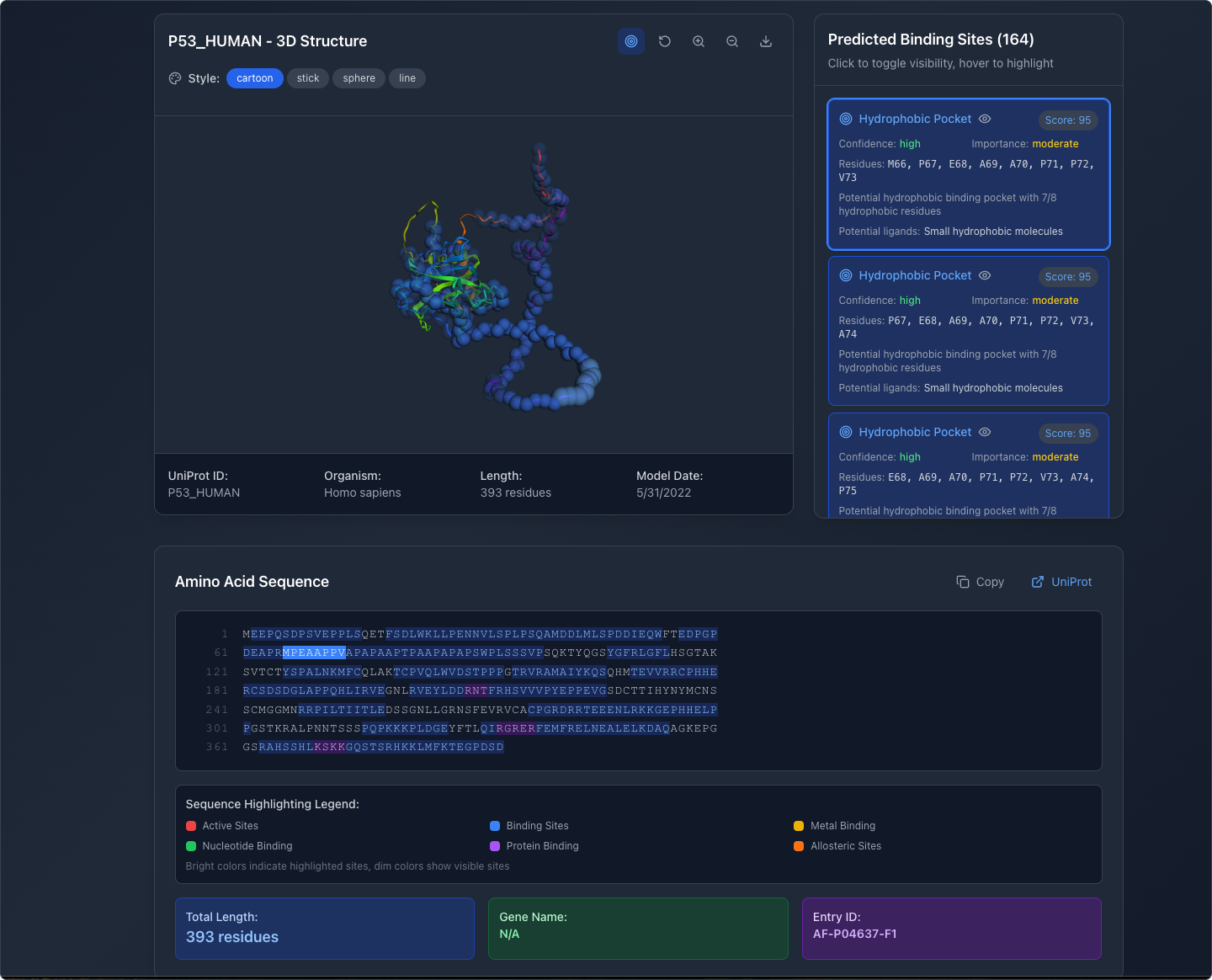
Load a Protein
The user enters a UniProt ID or amino acid sequence. The application retrieves the predicted structure from AlphaFold's database.
View Structure and Binding Sites
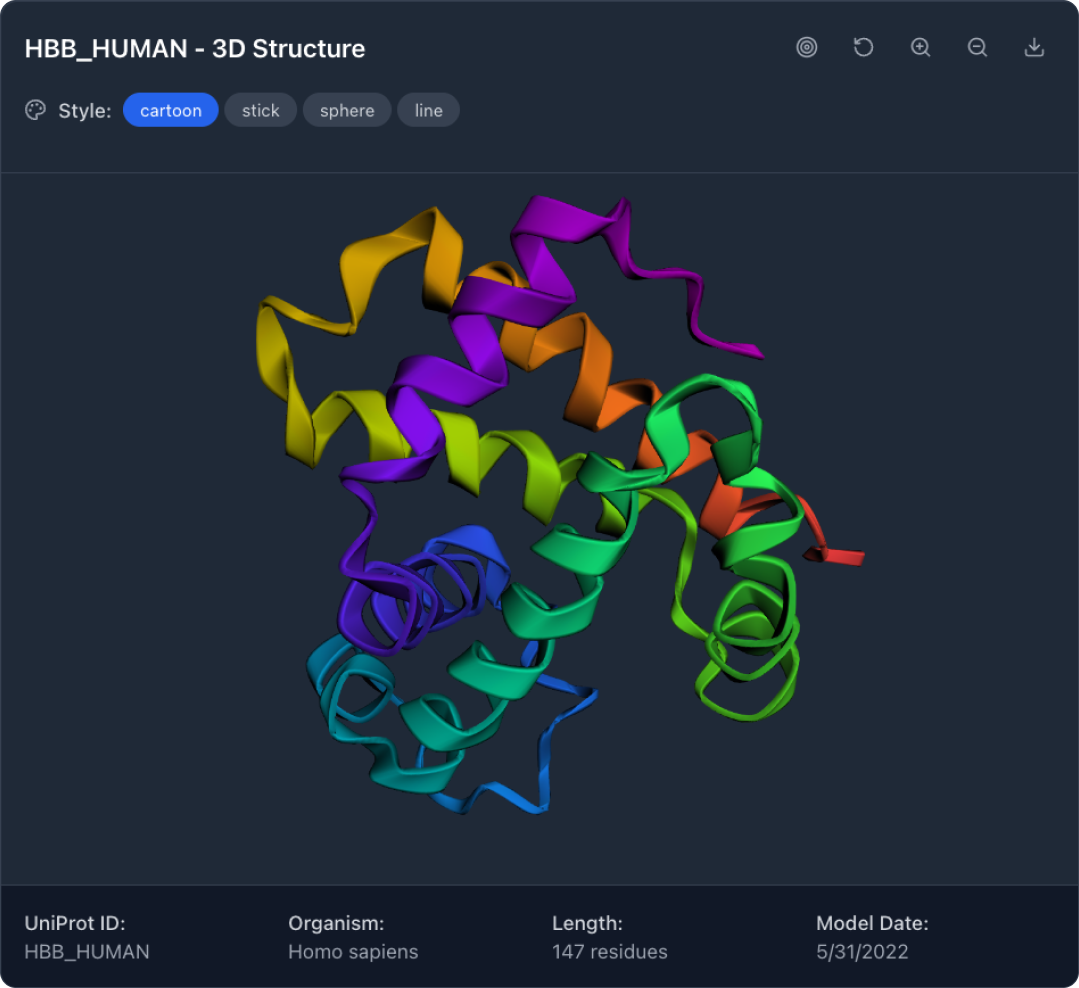
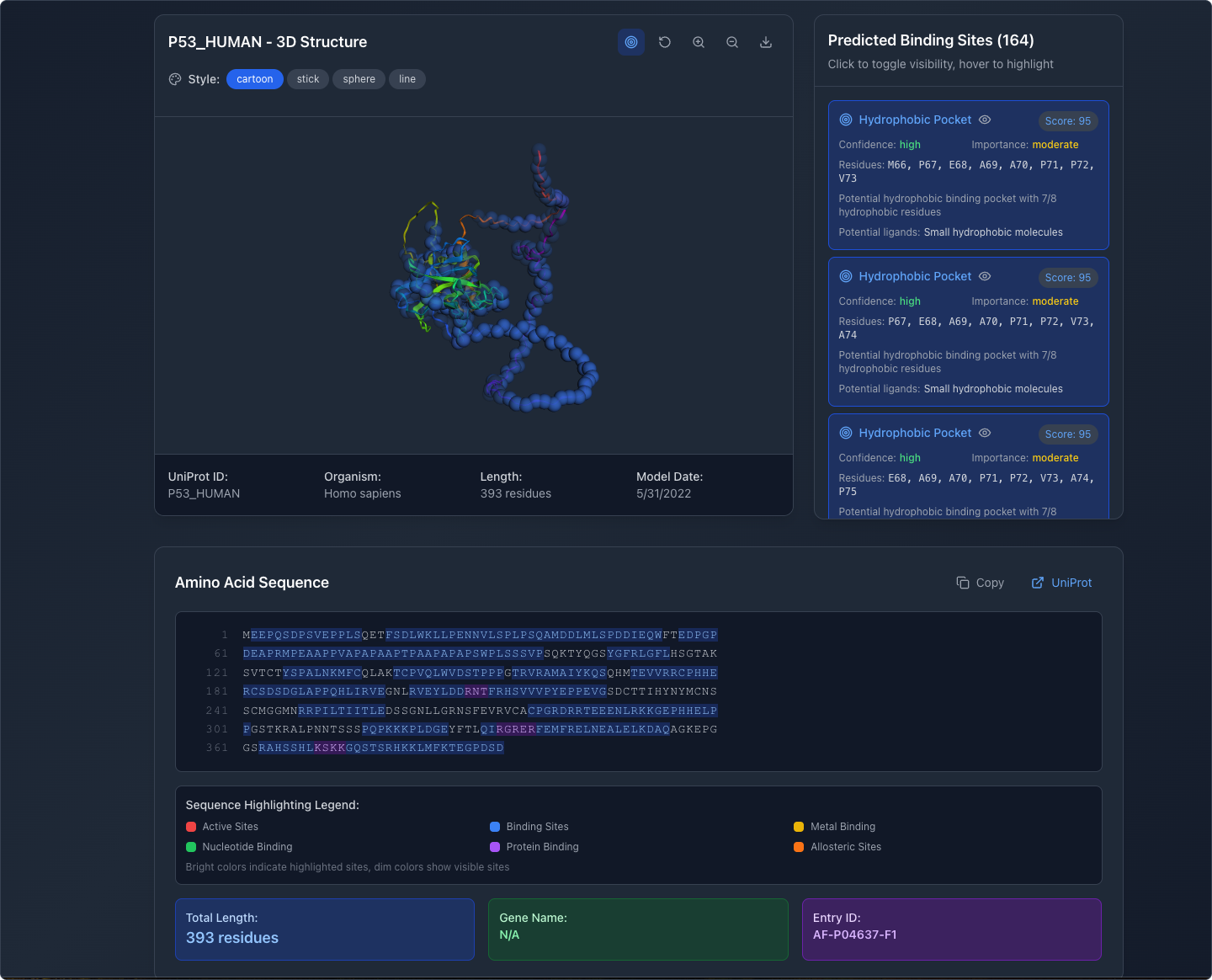
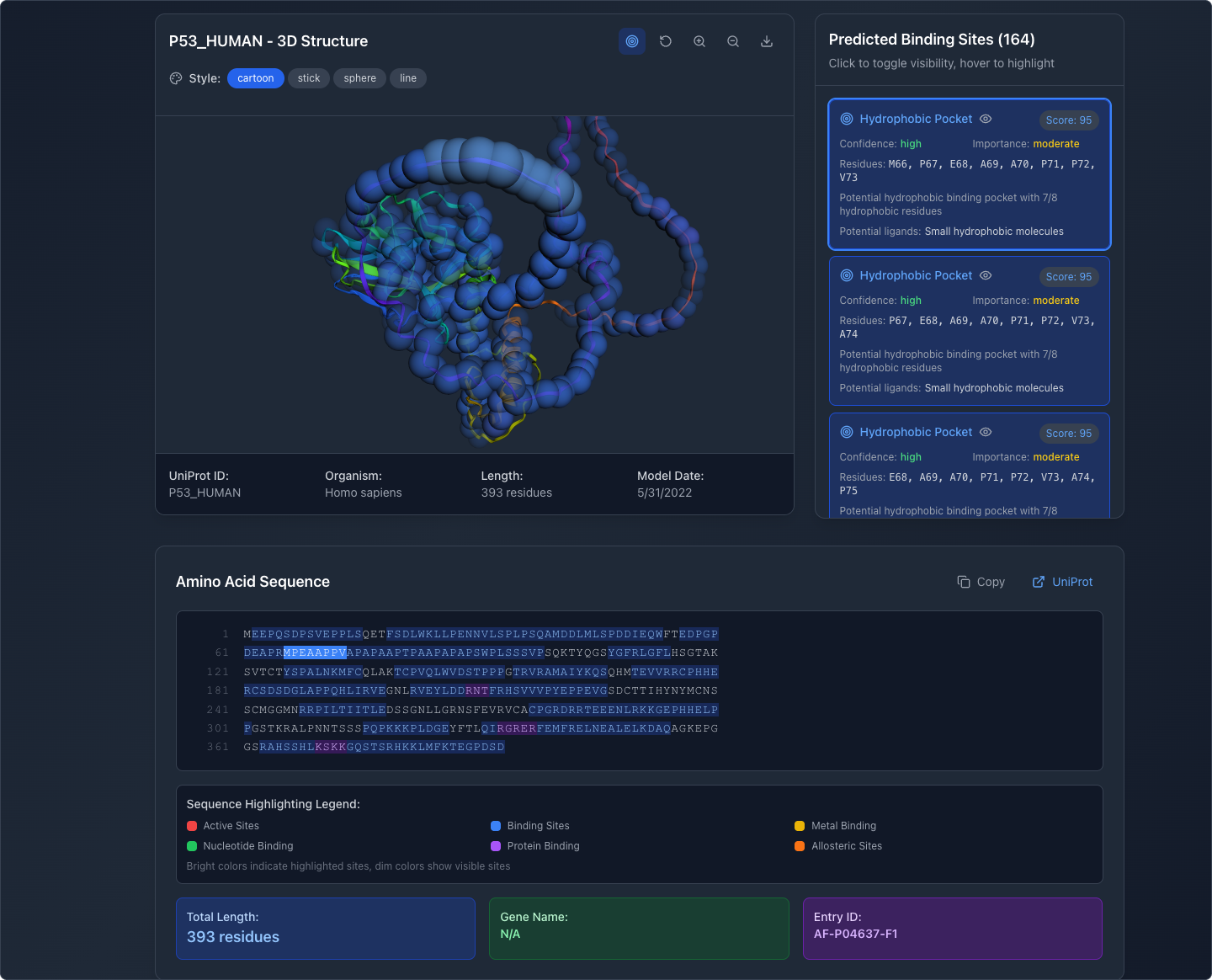
The app renders the protein's 3D structure and displays predicted binding sites as blue spheres. Users can explore the full molecule before selecting specific sites.
Inspect Individual Binding Sites
Selecting a binding site in the side panel highlights its position in the 3D viewport and in the amino acid sequence below, providing researchers with spatial and sequential context simultaneously.
Manipulate the View
Users can rotate, pan, and zoom the 3D model to examine structures from any angle. Display modes include cartoon, stick, sphere, and line representations, matching conventions familiar to researchers.
What This Demonstrates
This prototype is not a finished product. It's a proof of concept that demonstrates a specific capability: a designer with domain knowledge can use AI-assisted tools to build a functional, interactive application in a fraction of the time traditional methods would require.
In a real project, this kind of prototype becomes a powerful research and alignment tool. Instead of presenting stakeholders with static wireframes and asking them to imagine the interaction, you hand them something they can use. Feedback shifts from abstract ("I think the layout should be different") to concrete ("when I select this binding site, I need to see the neighboring residues too"). That changes the speed and quality of the entire product development process.
AI-assisted prototyping does not replace user research, information architecture, or interaction design. It accelerates the path from hypothesis to testable artifact, which means faster learning, better alignment, and less wasted effort.
This approach is now a standard part of how I work with product teams.